Unicycler

Unicycler is an assembly pipeline for bacterial genomes. It can assemble Illumina-only read sets where it functions as a SPAdes-optimiser. It can also assembly long-read-only sets (PacBio or Nanopore) where it runs a miniasm+Racon pipeline. For the best possible assemblies, give it both Illumina reads and long reads, and it will conduct a short-read-first hybrid assembly.
Requirements
- Linux or macOS
- Python 3.4 or later
- C++ compiler with C++14 support:
- setuptools (only required for installation of Unicycler)
- For short-read or hybrid assembly:
- SPAdes v3.14.0 or later (
spades.py)
- SPAdes v3.14.0 or later (
- For long-read or hybrid assembly:
- Racon (
racon)
- Racon (
- For rotating circular contigs:
- BLAST+ (
makeblastdbandtblastn)
- BLAST+ (
Unicycler expects external tools to be available in $PATH. If they aren't, you can specify their location using Unicycler options (e.g. --spades_path).
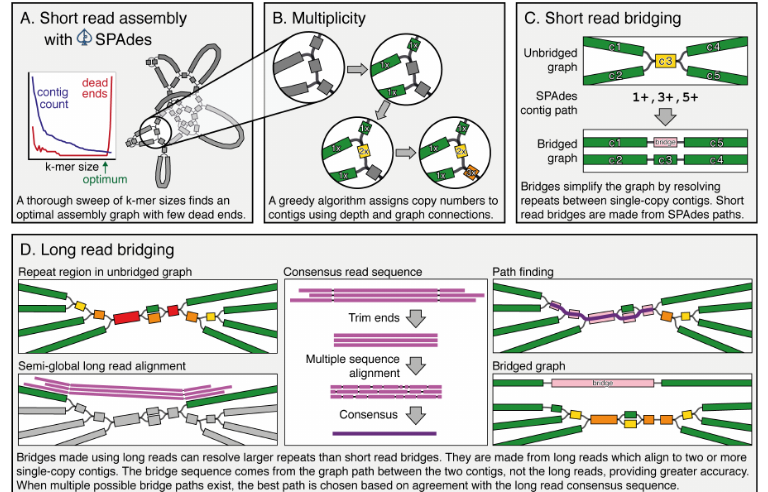
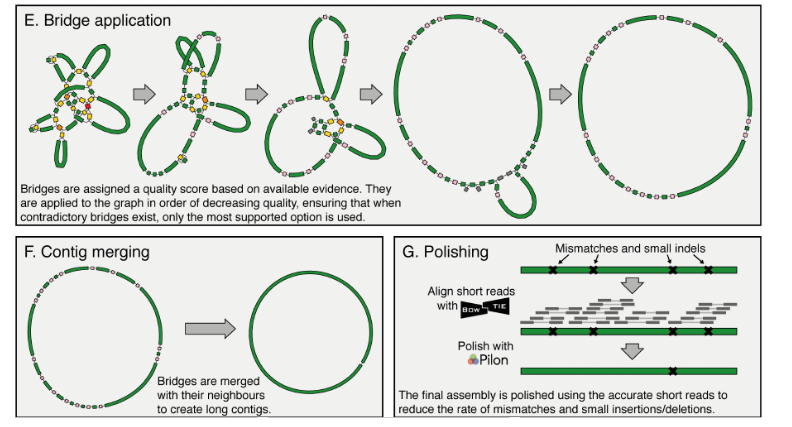
Figure 1: Key steps in the Unicycler pipeline steps in the Unicycler pipeline.