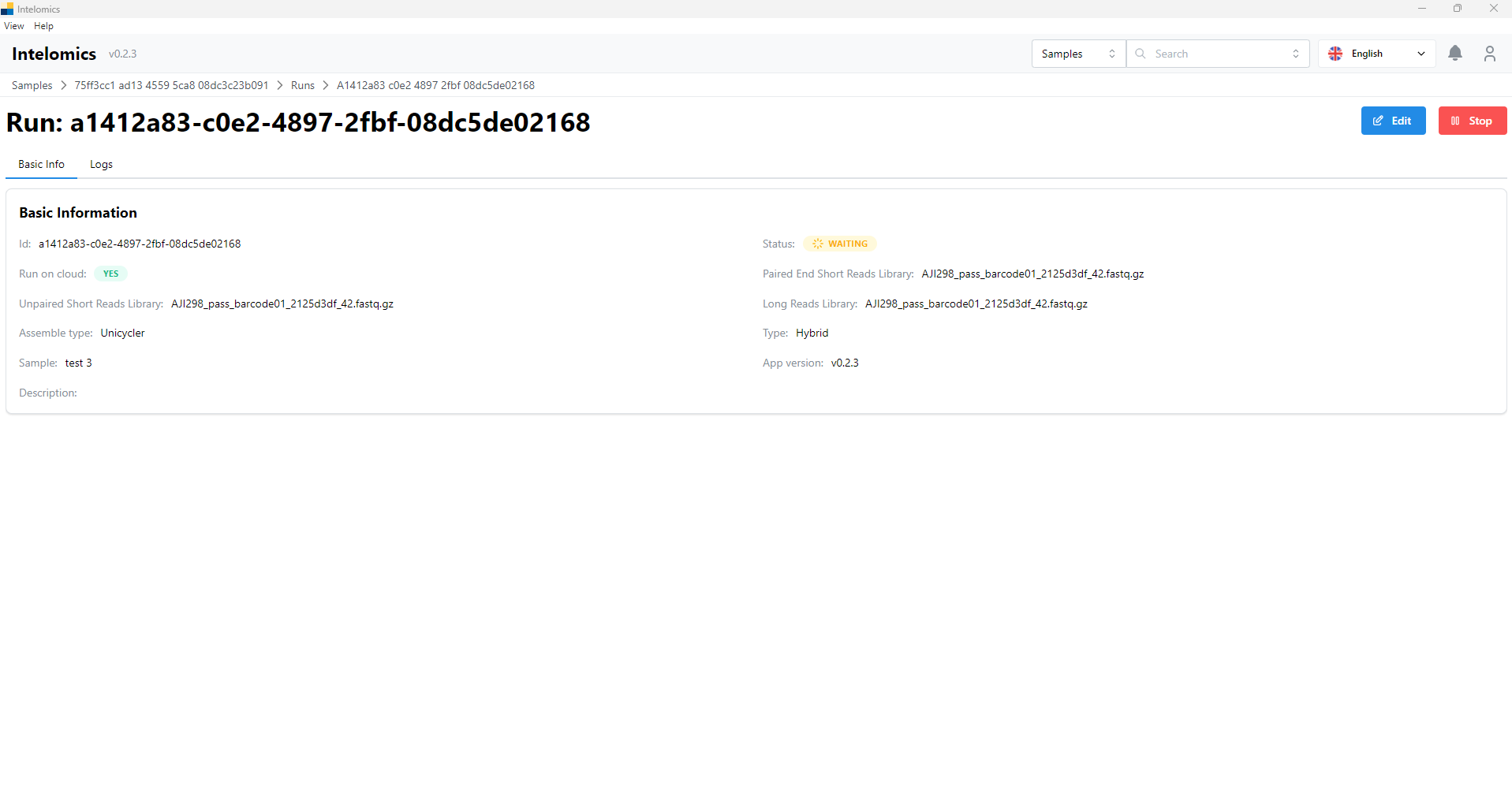
Run
Run Nanopore samples
- Users need to go to Samples:
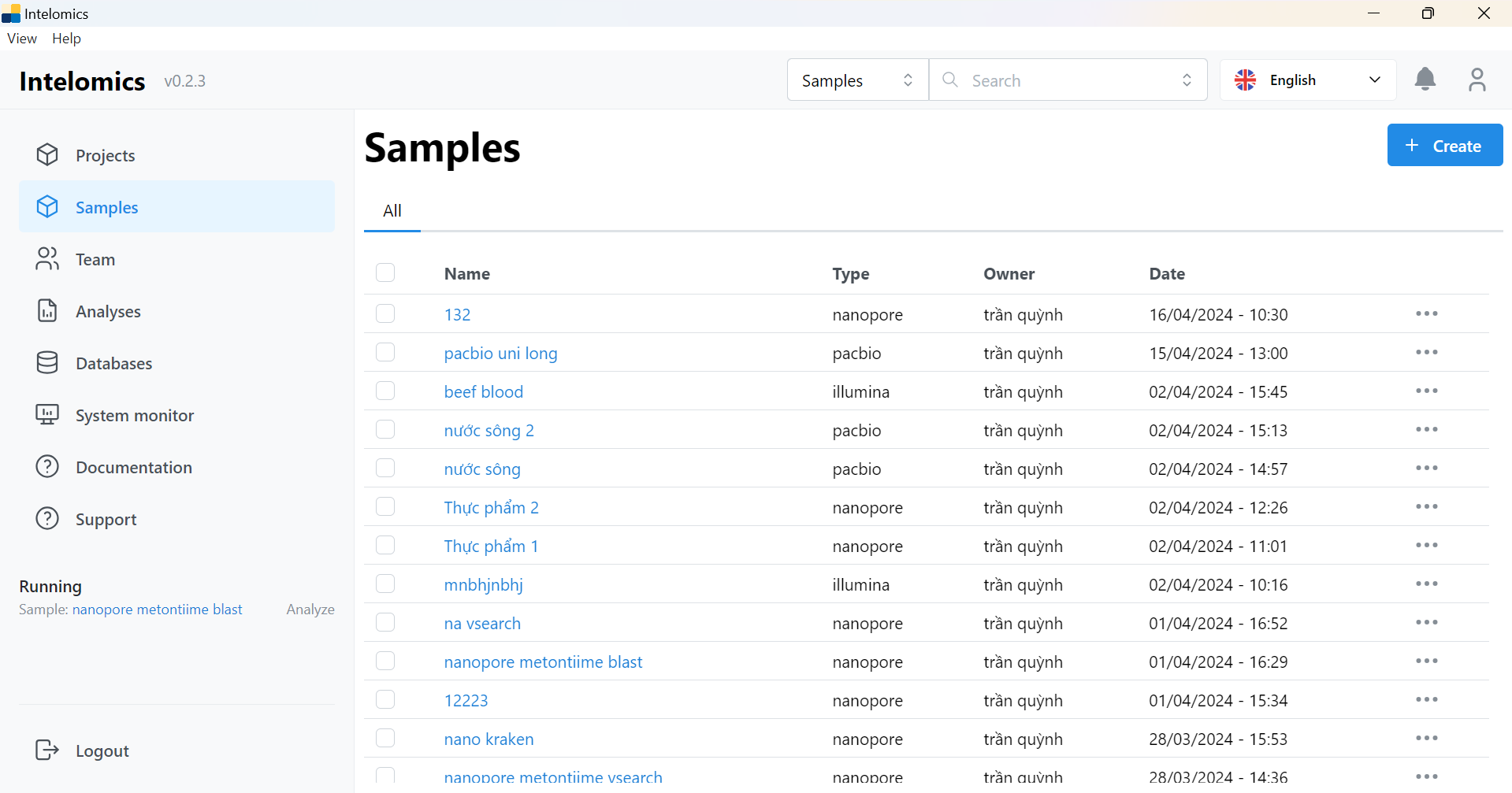
For example, in this case, the user needs to click on Name 123:
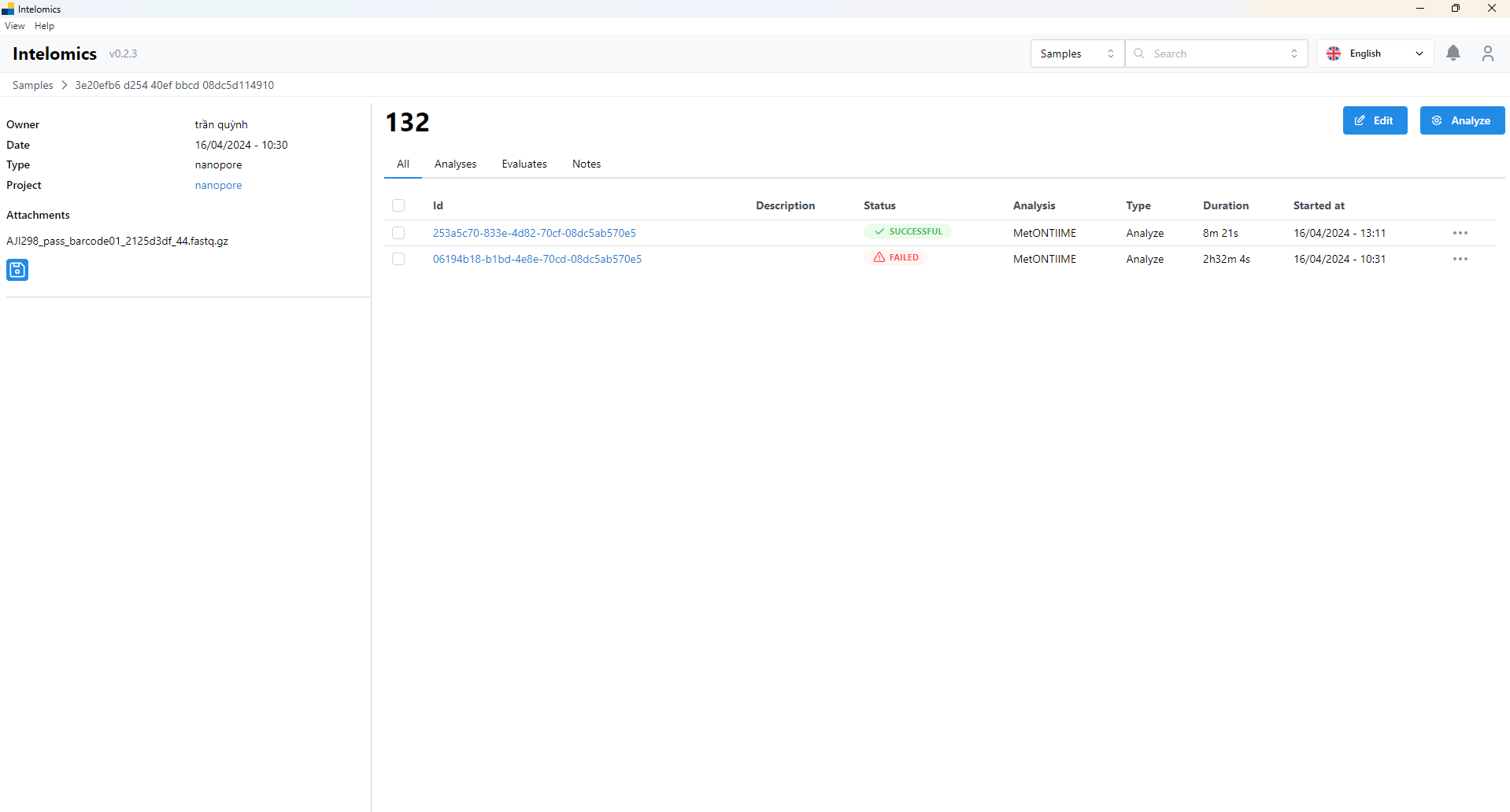
- Then the user continues to click on Id 253a5c70-833e-4d82-70cf-08dc5ab570e5

Here, the application will automatically download samples, then the user will receive:
- Basic Information
- Logs
- Result: contains a lot of information in tabs such as:
- demux_summary
- table
- rep-seqs
- taxonomy
- taxa-bar-plots
- taxa-bar-plots-no-Unassigned
- Taxonomy tree with read
- Result folder
-
If users want to Evaluate, please continue click Evaluate button, which is on the top-right of the screen. The interface of application will be:
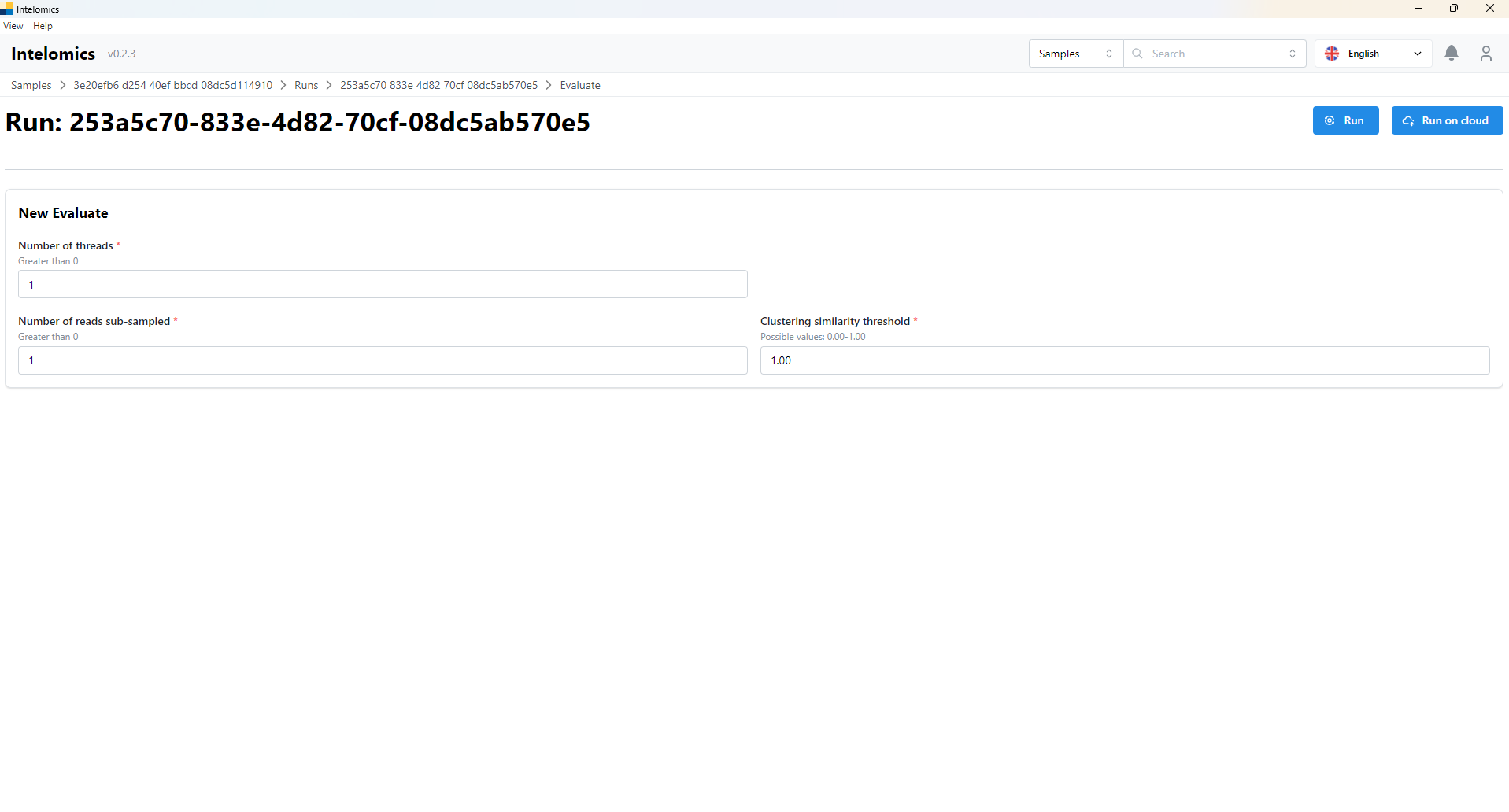
-
User have to input parameters into input box (Number of threads, Number of reads sub-sampled and Clustering similarity threshold)
-
User have 2 options: Run and Run on clound:
- If user option Run: It will run on your computer
- If user option Run on Cloud: It will run on cloud
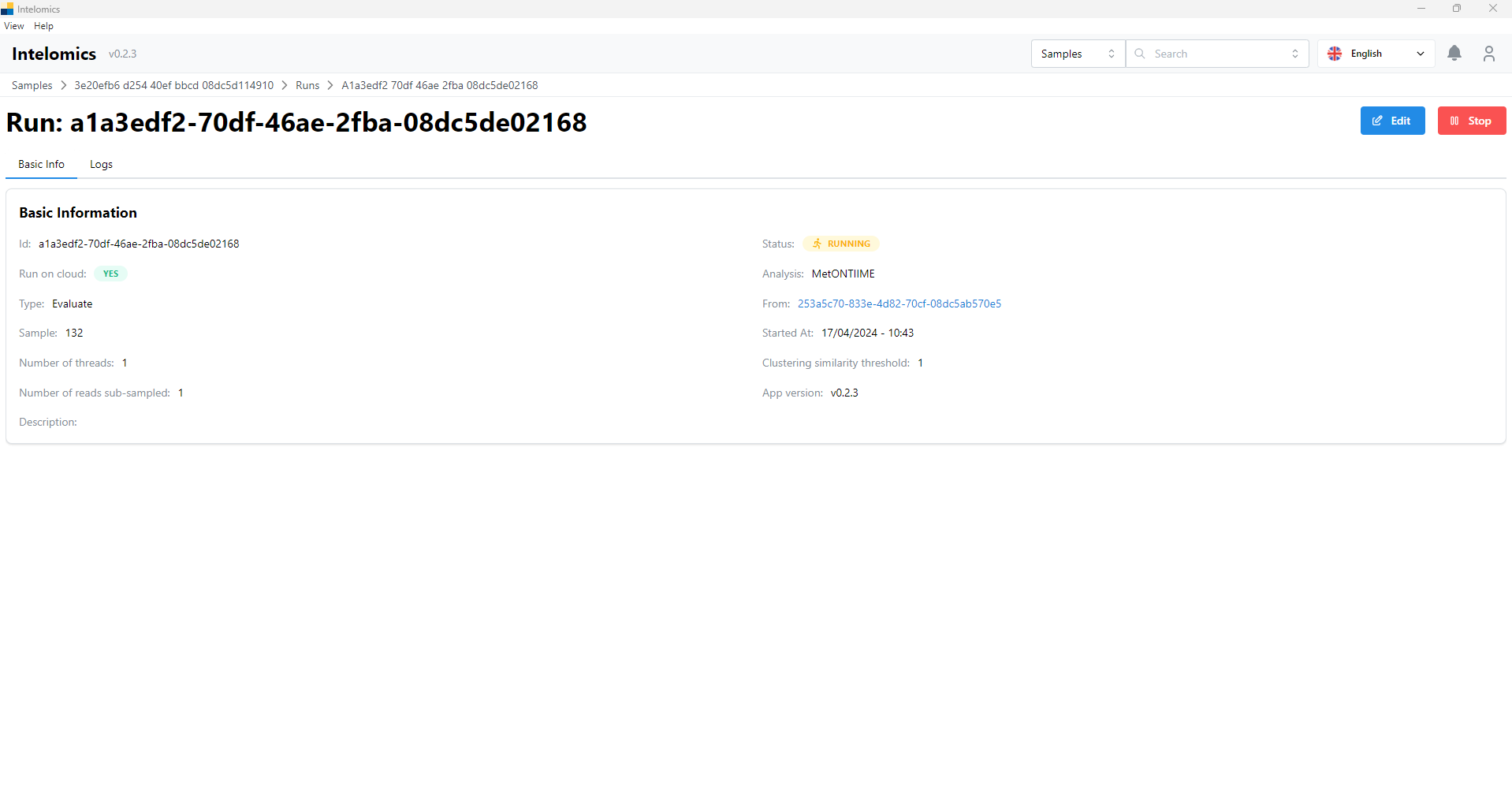
Moreover, when user click on Name 123, on the top-right of the screen, it have an Analyze button. When user click on this button, it will have:
User can option MetONTIIME or kraken2:
- If user option MetONTIIME: user have to input parameters into input box (Database sequence Database Taxonomy, Number of threads,...), then click Run button or Run on Cloud. It would be show basic information and logs
- If user option kraken2: user have to input parameters into input box (Database sequence), then click Run button or Run on Cloud. It would be show basic information and logs
Run Pacbio samples
- Users need to go to Samples:

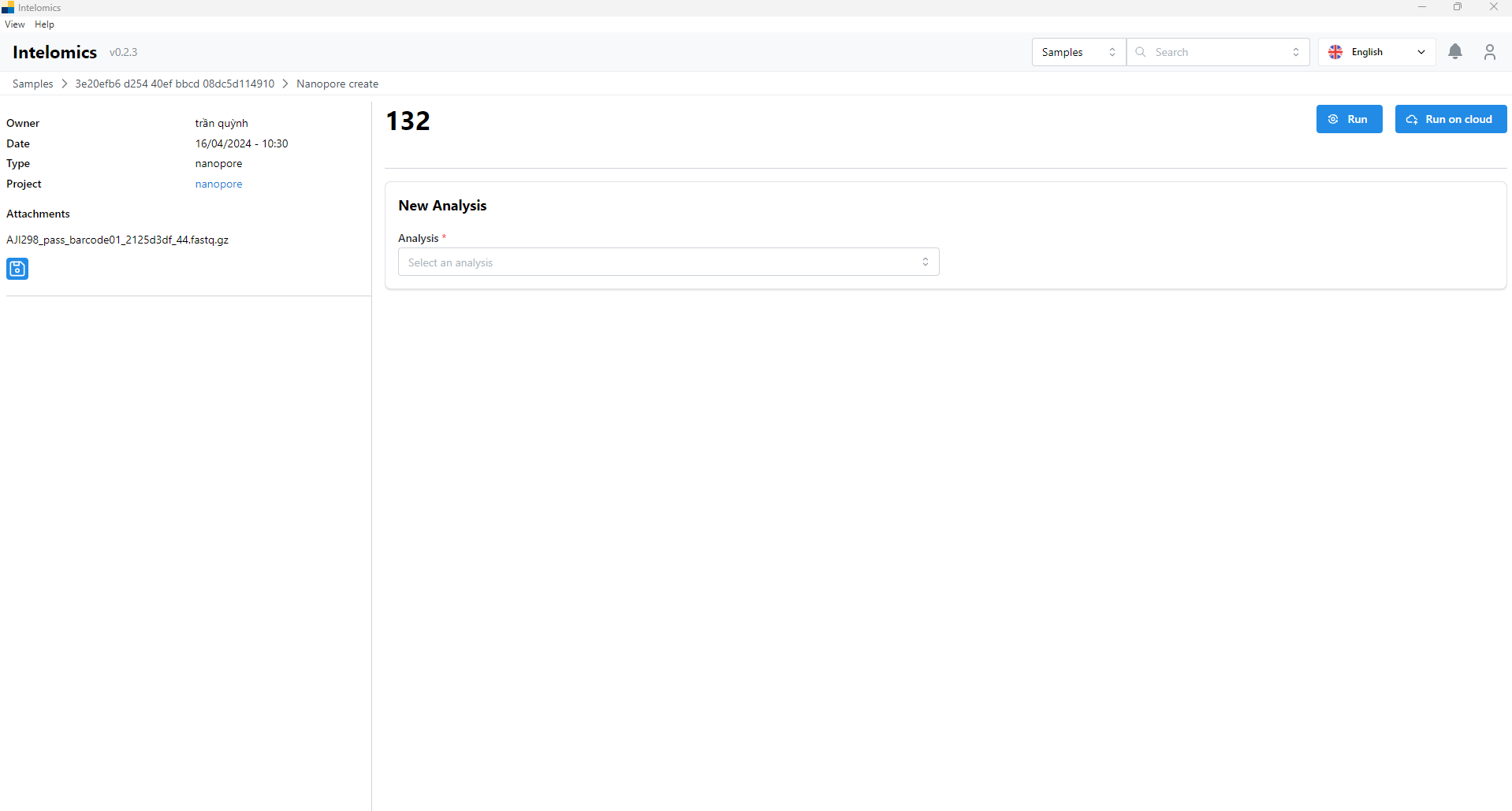
For example, in this case, the user needs to click on Name pacbio uni long:
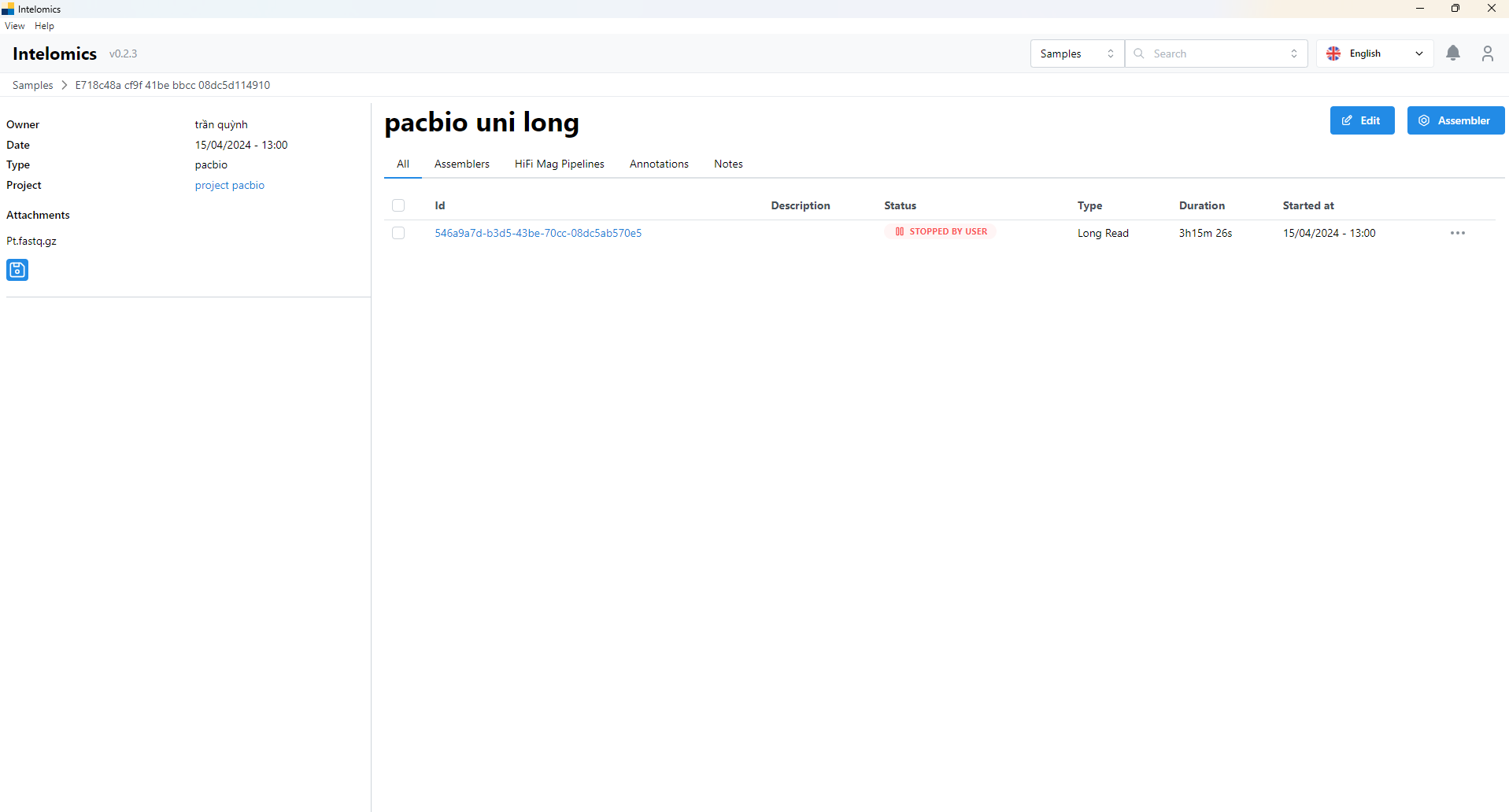
- Then the user continues to click on Id 546a9a7d-b3d5-43be-70cc-08dc5ab570e5
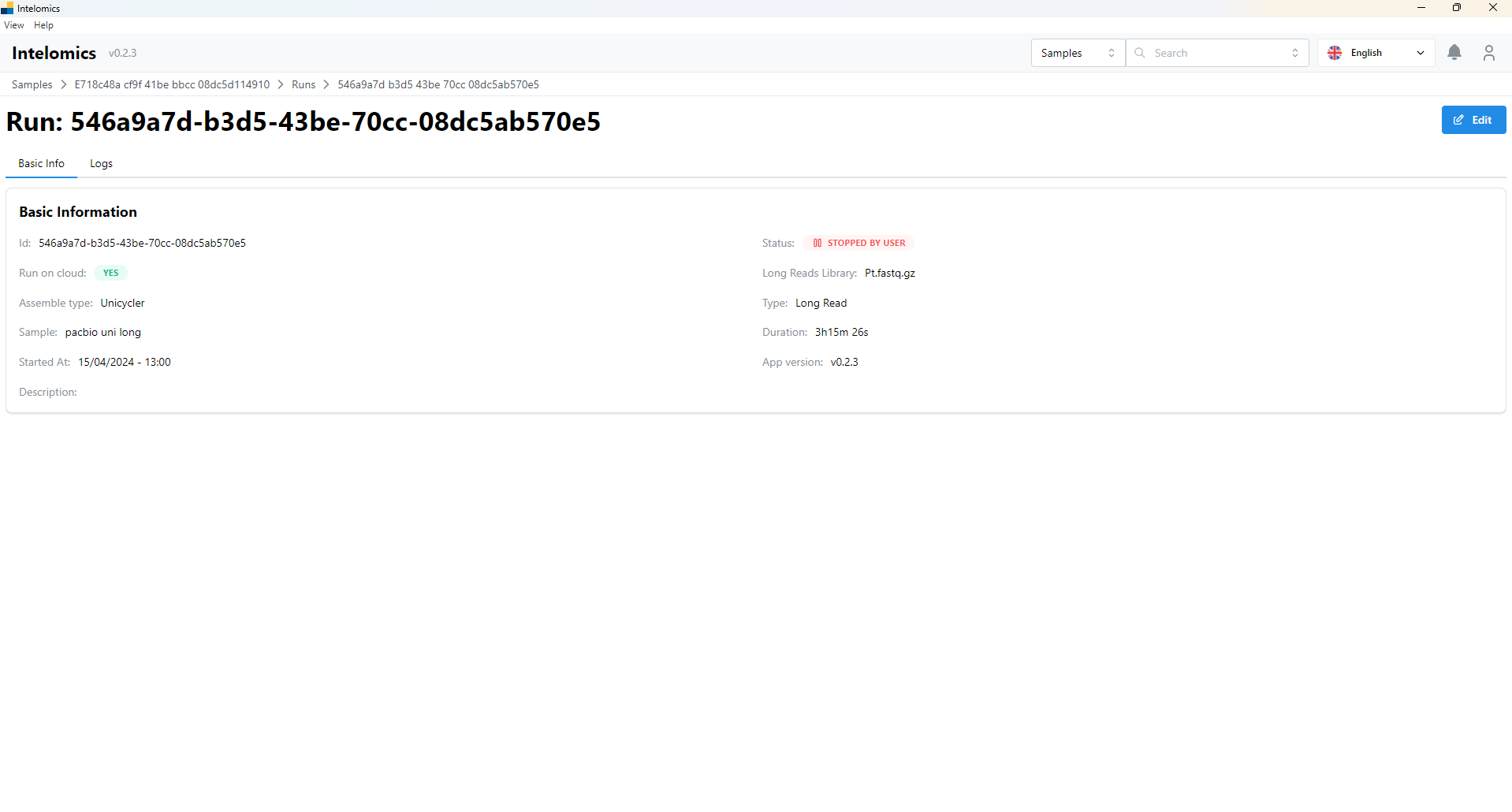
Here, the application will automatically download samples, then the user will receive:
- Basic Information
- Logs
User can choose Hifi Mag Pipeline button or Hifi Mag Pipeline on Cloud. User would be recieved:
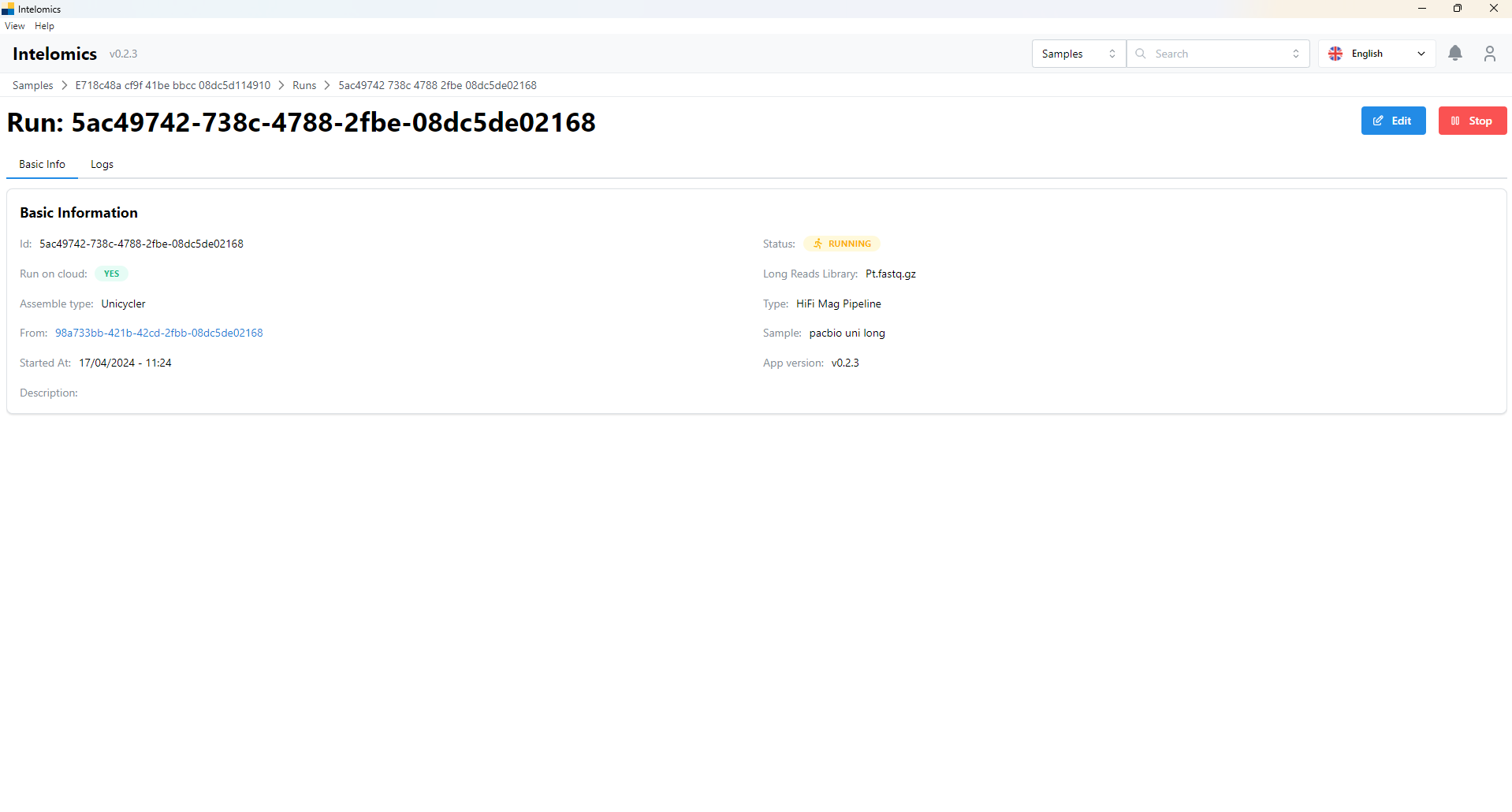
Moreover, when user click on Name pacbio uni long, on the top-right of the screen, it have an Assembler button. When user click on this button, it will have:
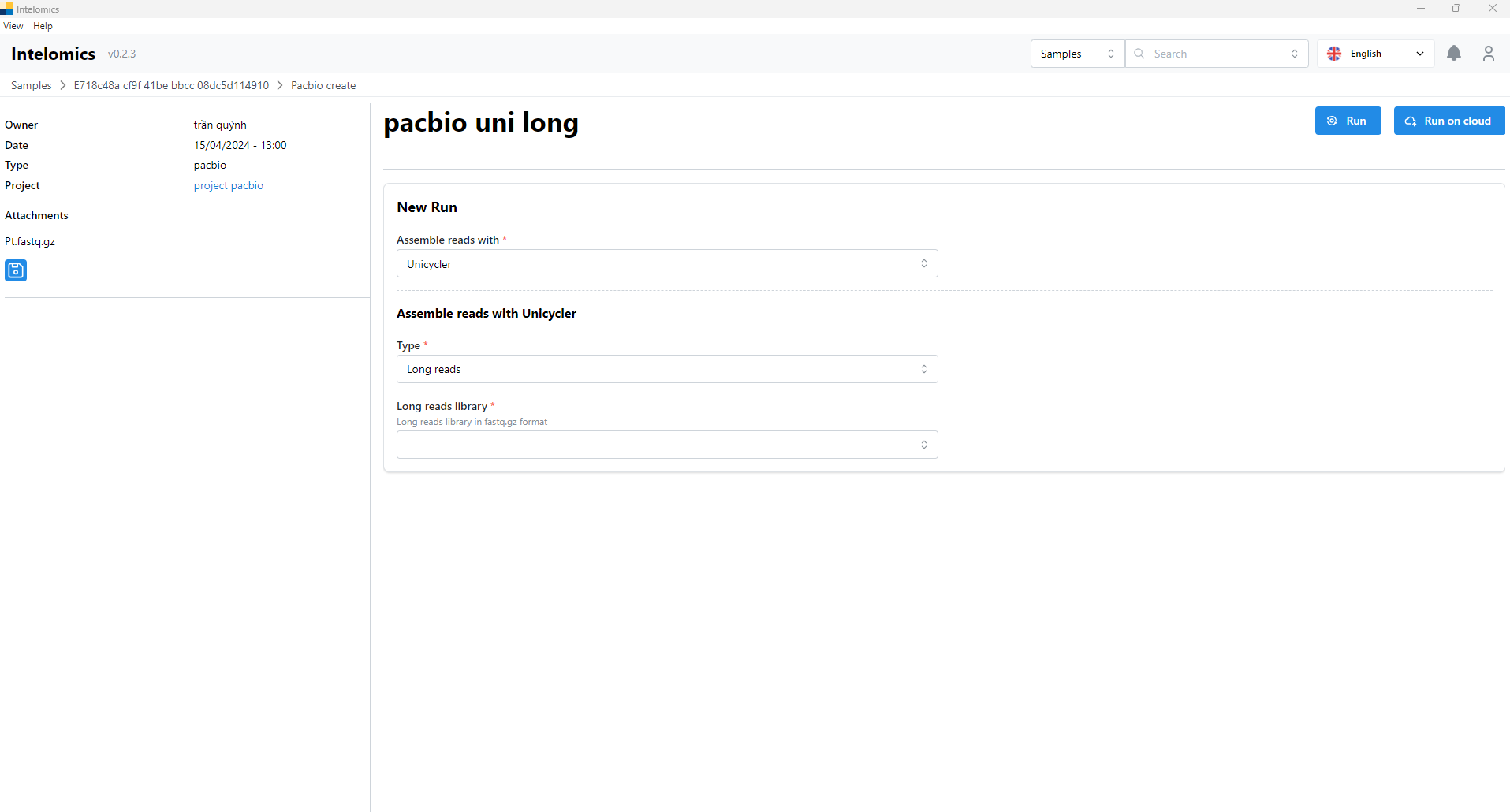
User need to select options of input box, then user can click Run button or Run on clound. The result will be:

Run Illumina samples
- Users need to go to Samples:

For example, in this case, the user needs to click on Name beef blood:
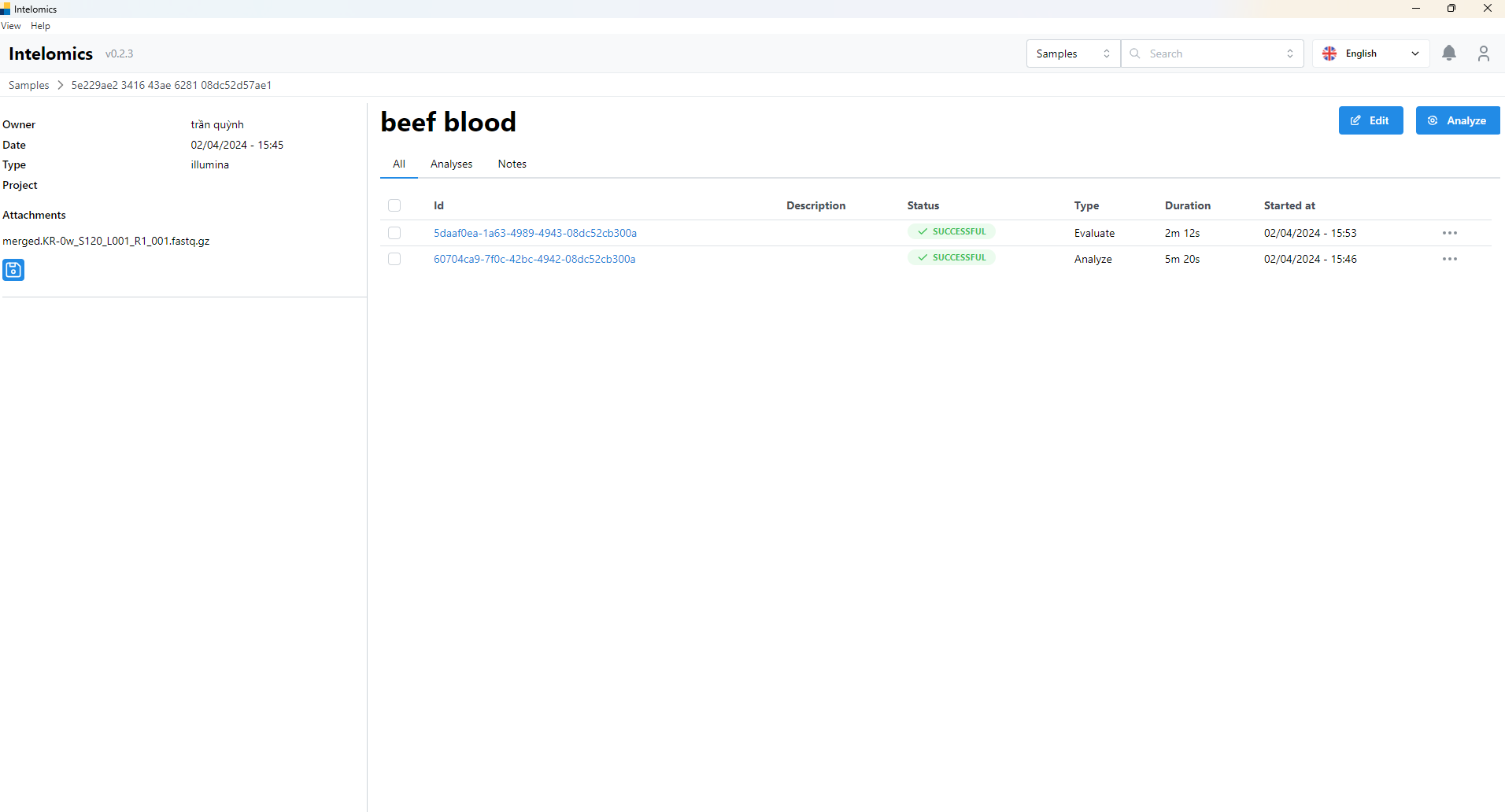
- Then the user continues to click on Id 5daaf0ea-1a63-4989-4943-08dc52cb300a
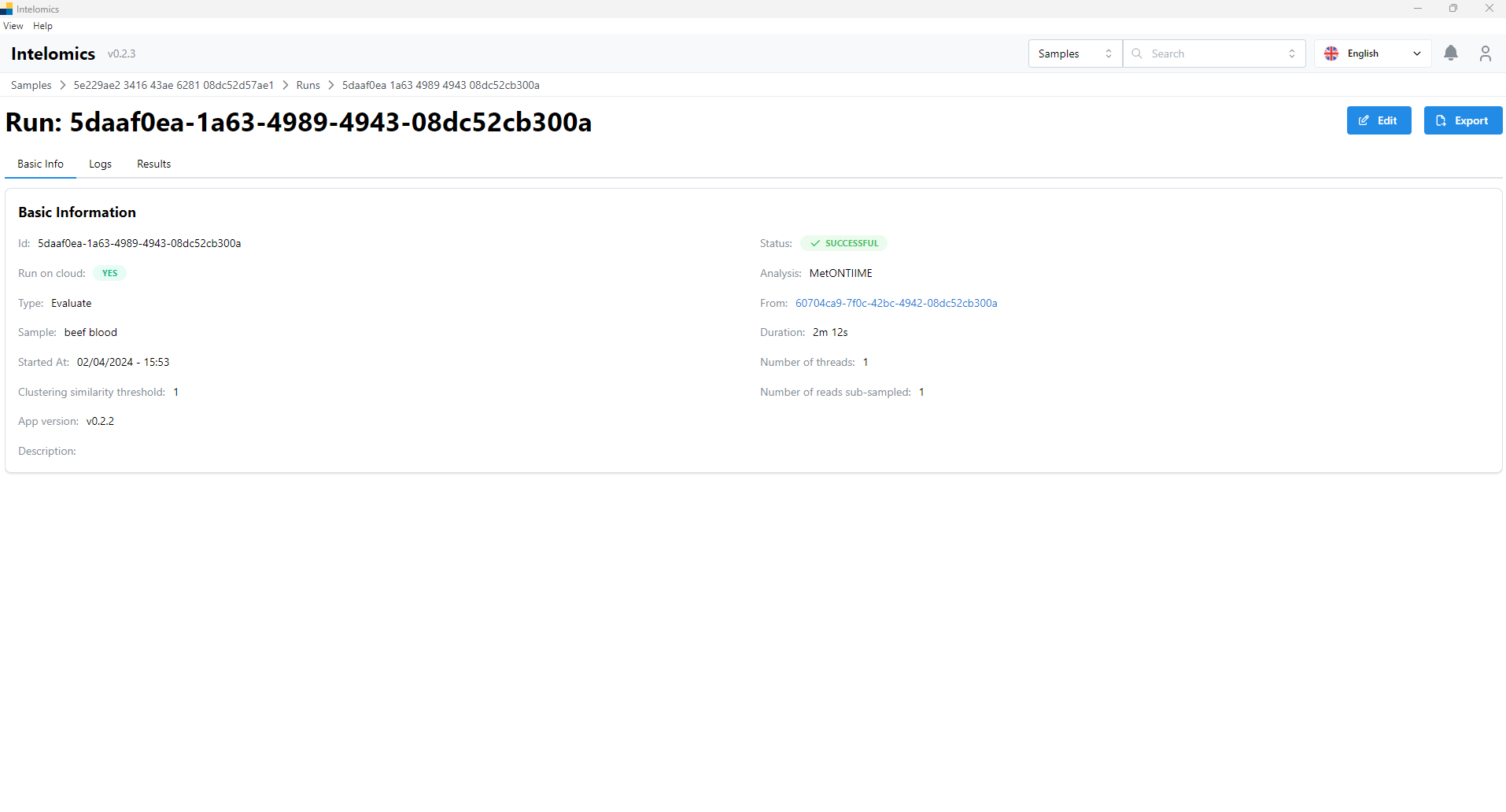
Here, the application will automatically download samples, then the user will receive:
-
Basic Information
-
Logs
-
Result: contains a lot of information in tabs such as:
- demux_summary
- table
- rep-seqs
- taxonomy
- taxa-bar-plots
- taxa-bar-plots-no-Unassigned
- Taxonomy tree with read
- Result folder
Moreover, when user click on Name 123, on the top-right of the screen, it have an Analyze button. When user click on this button, it will have:
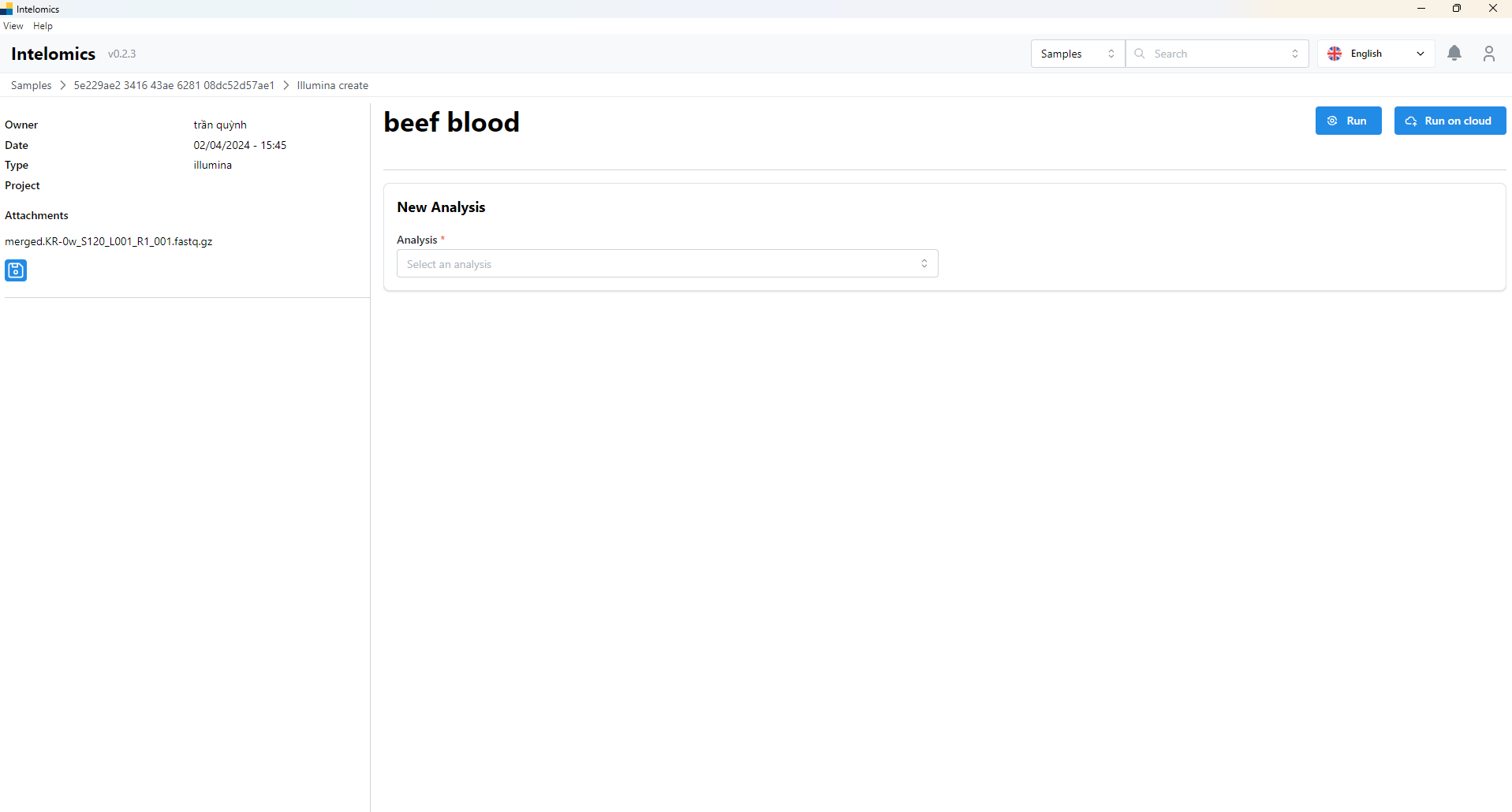
User can option MetONTIIME or kraken2:
- If user option MetONTIIME: user have to input parameters into input box (Database sequence Database Taxonomy, Number of threads,...), then click Run button or Run on Cloud. It would be show basic information and logs
- If user option kraken2: user have to input parameters into input box (Database sequence), then click Run button or Run on Cloud. It would be show basic information and logs
Run dbnseq samples
- Users need to go to Samples:

For example, in this case, the user needs to click on Name test3:
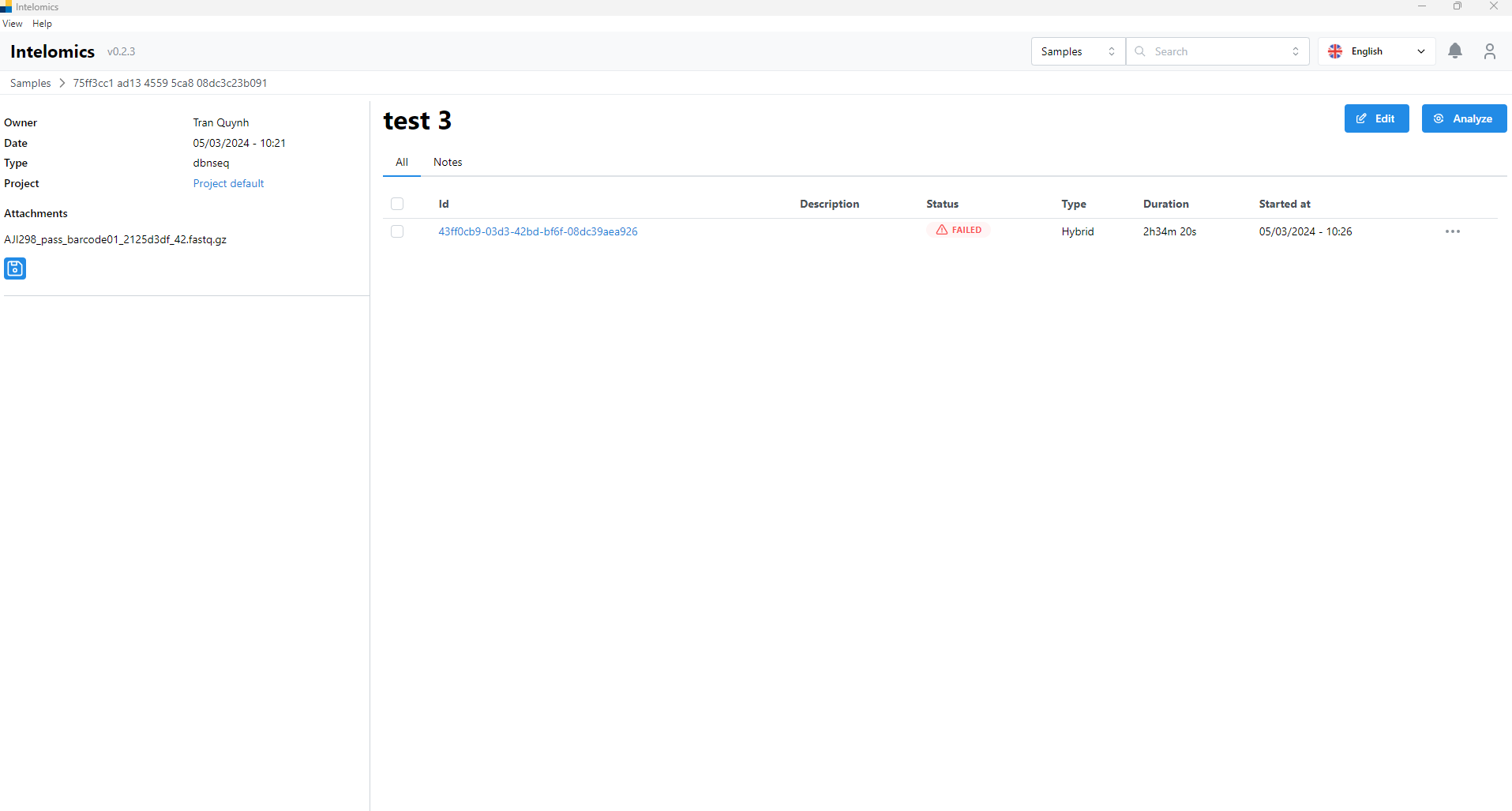
- Then the user continues to click on Id 43ff0cb9-03d3-42bd-bf6f-08dc39aea926
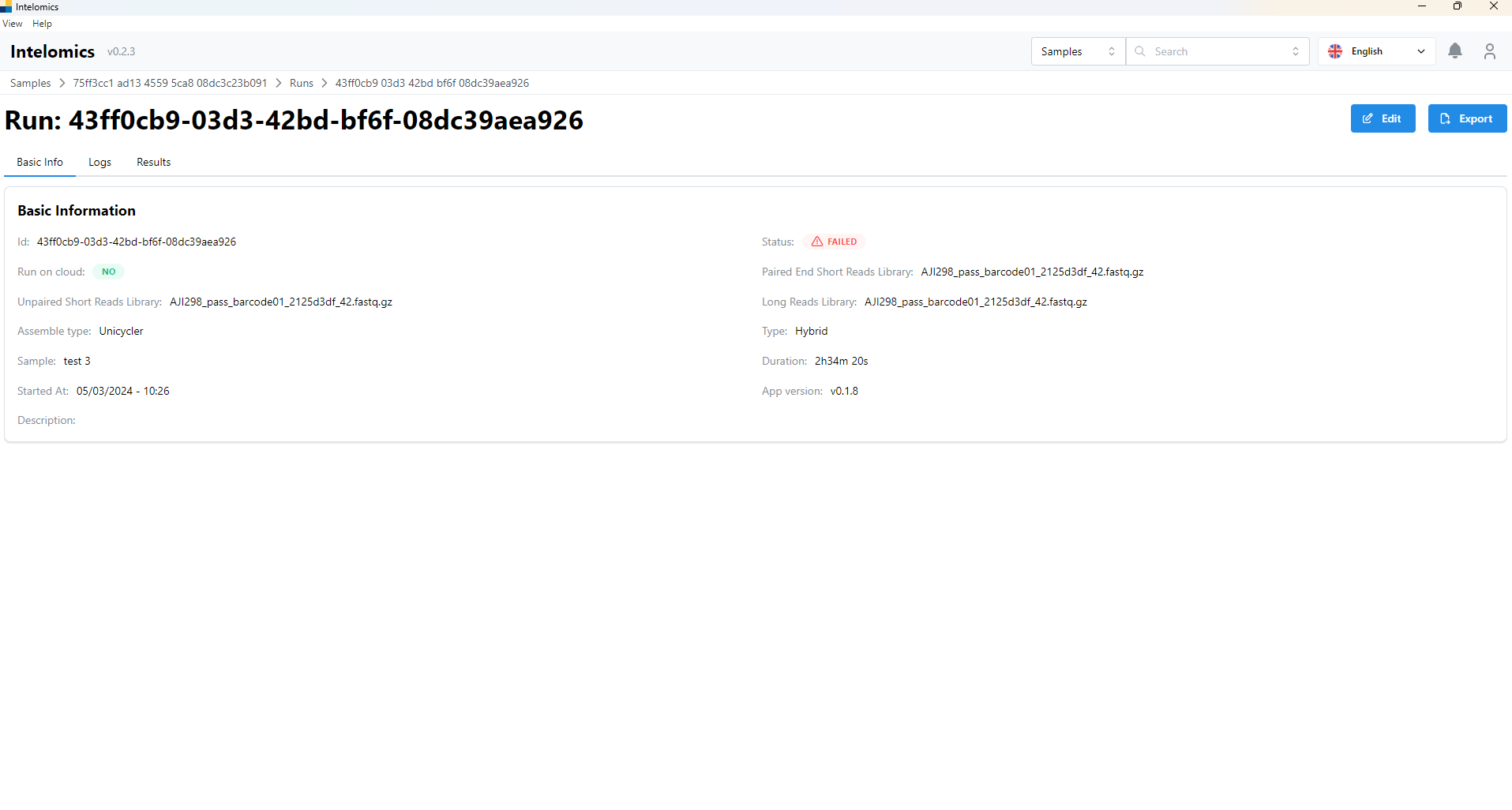
Here, the application will automatically download samples, then the user will receive:
-
Basic Information
-
Logs
-
Result: contains a lot of information in tabs such as:
- demux_summary
- table
- rep-seqs
- taxonomy
- taxa-bar-plots
- taxa-bar-plots-no-Unassigned
- Taxonomy tree with read
- Result folder
Moreover, when user click on Name 123, on the top-right of the screen, it have an Analyze button. When user click on this button, it will have:
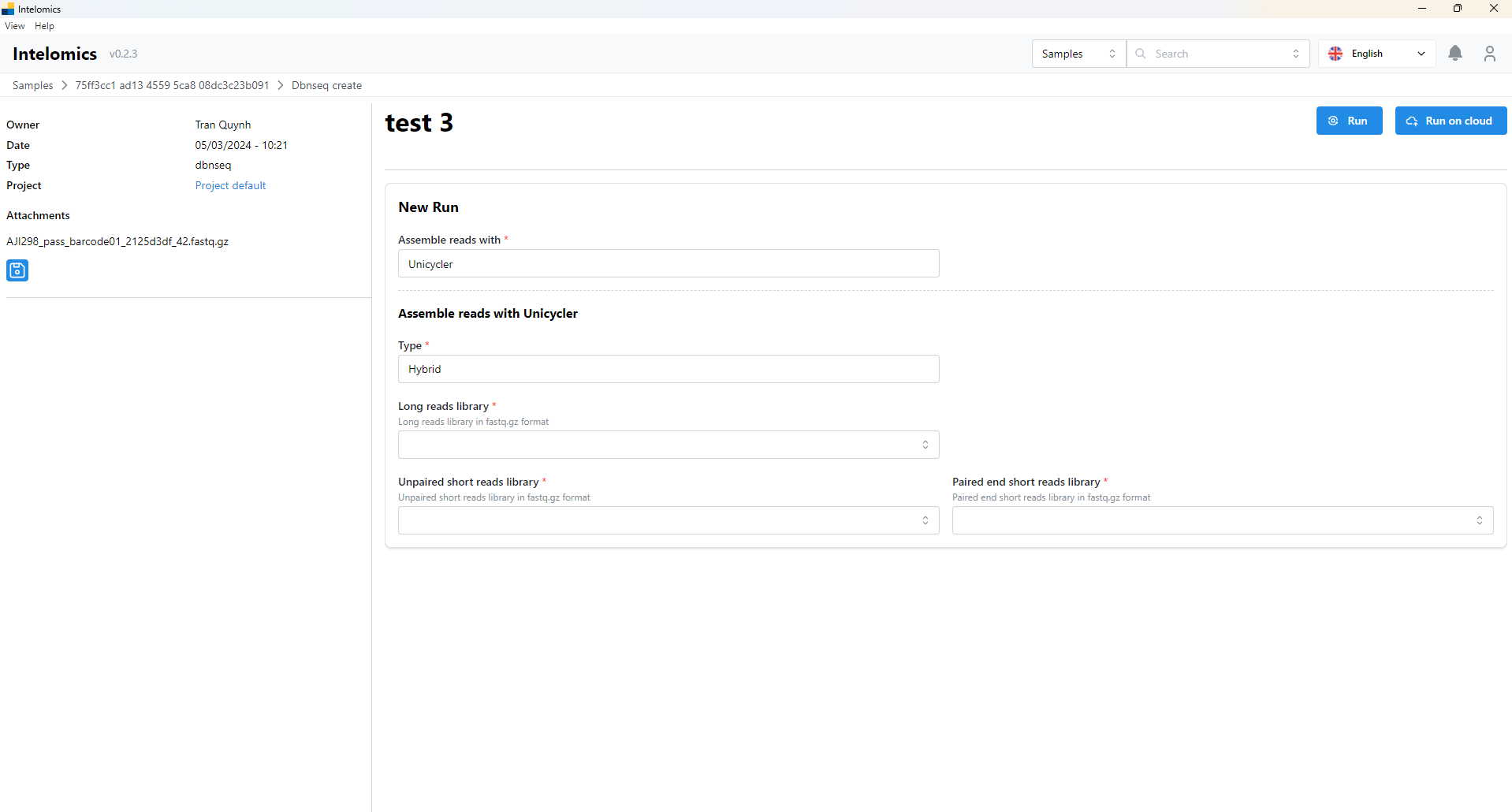
You have to select options of input box (Assemble reads with, Type, Long reads library ,..) then click Run button or Run on Cloud. It would be show basic information and logs: